Using Julia on EDDIE at University of Edinburgh
TL;DR :
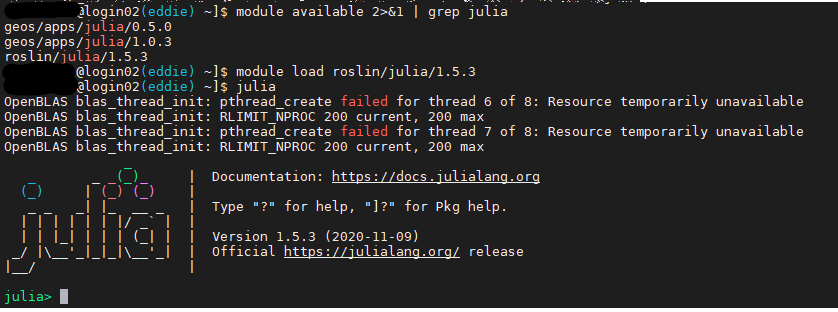
module load roslin/julia/1.5.3
For my next project during my PhD, I’ve decide to use the programming language Julia. Although it is much faster than Python, my usual language, there are still a few simulation I want to run that will take a lot of time. Previously I’ve used EDDIE, the HPC cluster at the University of Edinburgh when I needed a little more computing power, or when I expect simulations to take a while and don’t want to brick my laptop. MATLAB and Python usage is well documented on the UoE wiki, but now I need to work out how to get Julia going. This process will work for any language you want to see if is available on the cluster.
As usual, first of all you’ll have to sign into the UoE VPN, and ssh into the cluster. These steps will be familiar if you’ve ever logged into the cluster before, and are well documented online. To find out whether the module exists, we can use module available.
This spits out a whole load of text showing everything available on the cluster.
First up we see our old friends MATLAB, Python and R.
 Scrolling further down we see that Julia is listed in a folder belonging to the Roslin Institute. The most obvious thing to try here is
Scrolling further down we see that Julia is listed in a folder belonging to the Roslin Institute. The most obvious thing to try here is module load julia, but that won’t work. Instead we have to be more specific on the path, using
module load roslin/julia/1.5.3
This is the most recent version available. Alternatively to sifting through all available modules ourselves, we can use
module available 2>&1 | grep julia
and this will do the hard work for us, listing only the modules with names including Julia.
After running the load command, we can start up a Julia REPL as normal by just typing julia. To use Julia non-interactively, use the line above to load Julia as part of your job script and then julia my_script.jl. This can be submitted as usual using qsub.
